Research
Mechanisms of Response and Resistance in Acute Myeloid Leukemias Characterized by Hyperactive Ras Signaling and Fusion Oncogenes
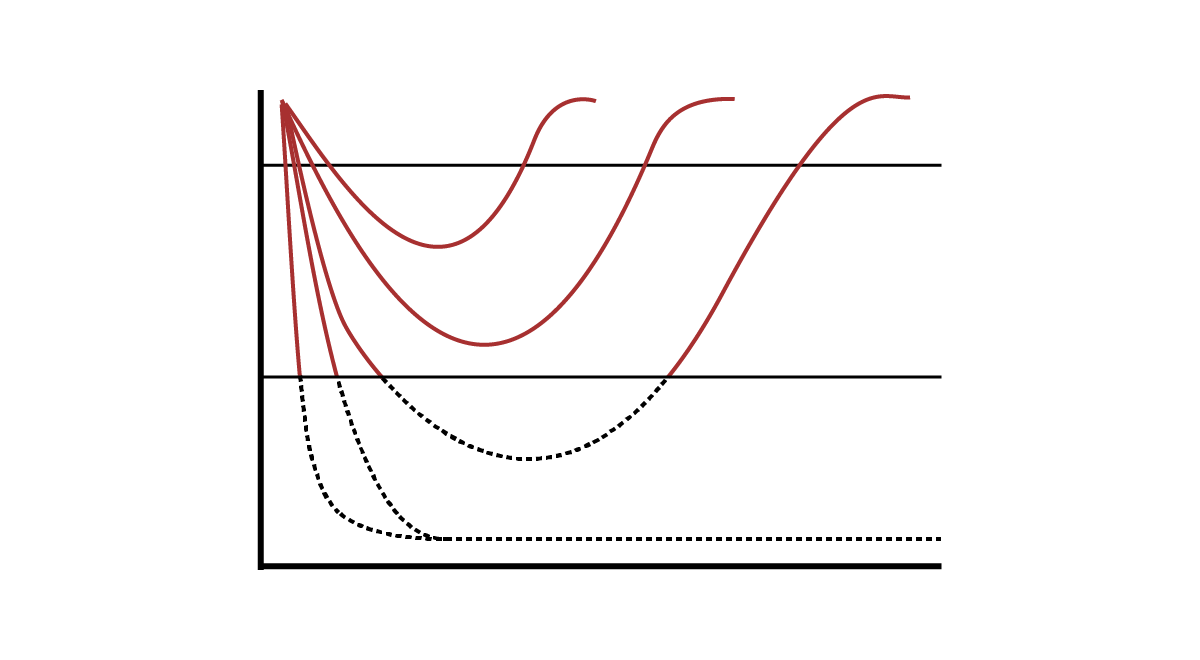
Somatic NRAS and KRAS mutations or NF1 inactivation occur in 20-25% of acute myeloid leukemias (AMLs) and are particularly common in children, adolescents, and young adults diagnosed with AML. In addition, RAS mutations are frequently present at relapse after responses to FLT3-ITD inhibitors, IDH1/2 inhibitors, and venetoclax. We harness accurate primary mouse models of AMLs characterized by hyperactive Ras signaling. We treat cohorts of recipient mice transplanted with AMLs in vivo with promising inhibitors targeting Ras or downstream effector pathways alone and in combination with chemotherapy or other promising targeted therapies. A subset of these AMLs respond dramatically to different treatments in vivo, but ultimately relapse and acquire drug resistance on the basis of in vivo validation. We are characterizing mechanisms of drug resistance, identifying pathways that modulate sensitivity to individual agents and drug combinations, and extending this work to human patient derived xenograft (PDX) models of AML.
Fusion oncogenes occur in 50-60% of AMLs diagnosed in children, adolescent, and young adults. Menin is an essential transcriptional cofactor for the initiation and maintenance of KMT2A-rearranged leukemias and menin inhibitors have shown promise in early phase clinical trials. We are investigating candidate mechanisms of de novo resistance and our recent data reveal that TP53 mutations (leading to inactivation of p53) confer resistance to the menin inhibitor revumenib. These data have implications for identifying AML patients who are at high risk of failing menin inhibitor treatment and for developing therapeutic strategies for overcoming resistance.
Transcriptional Plasticity in Leukemia Persistence Cells
Our preclinical therapeutic studies (and related genomic studies) have revealed candidate transcriptional biomarkers and mechanisms of drug resistance. Understanding how to more accurately predict leukemia relapse and how to more effectively treat persistent leukemia cells remain key barriers to improving outcomes in AML. Specifically, our preliminary data reveals that genes associated with upregulated expression in leukemia stem cells (LSCs) or initiating cells (LICs) can be harnessed to improve relapse risk prediction in pediatric AML. Concurrently, our preclinical therapeutic data implicates lineage plasticity playing a central role in drug resistance and the survival of leukemia persistence cells (LPCs). While these preliminary LIC and LPC studies have shown promise, they are also limited by bulk cell population analysis. Thus, we are leveraging high-resolution single cell platforms to investigate LIC and LPC populations, with the goal to improve relapse risk prediction and overcome leukemia persistence in pediatric AML.
Targeting Oncogenic Ras in Cancer
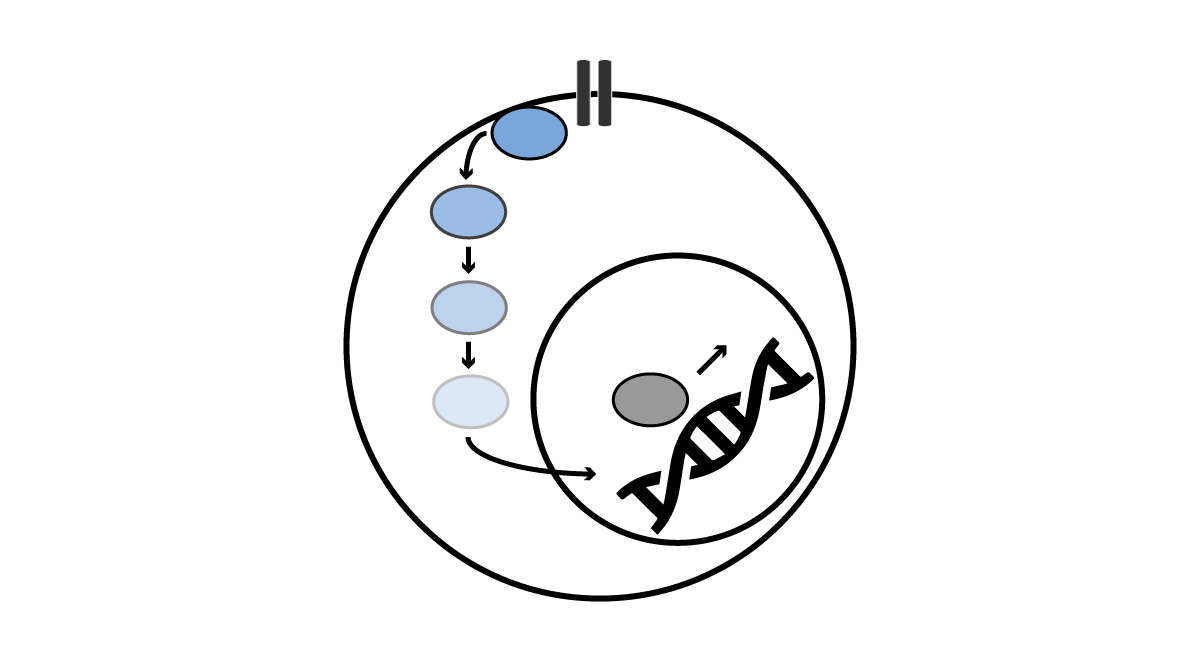
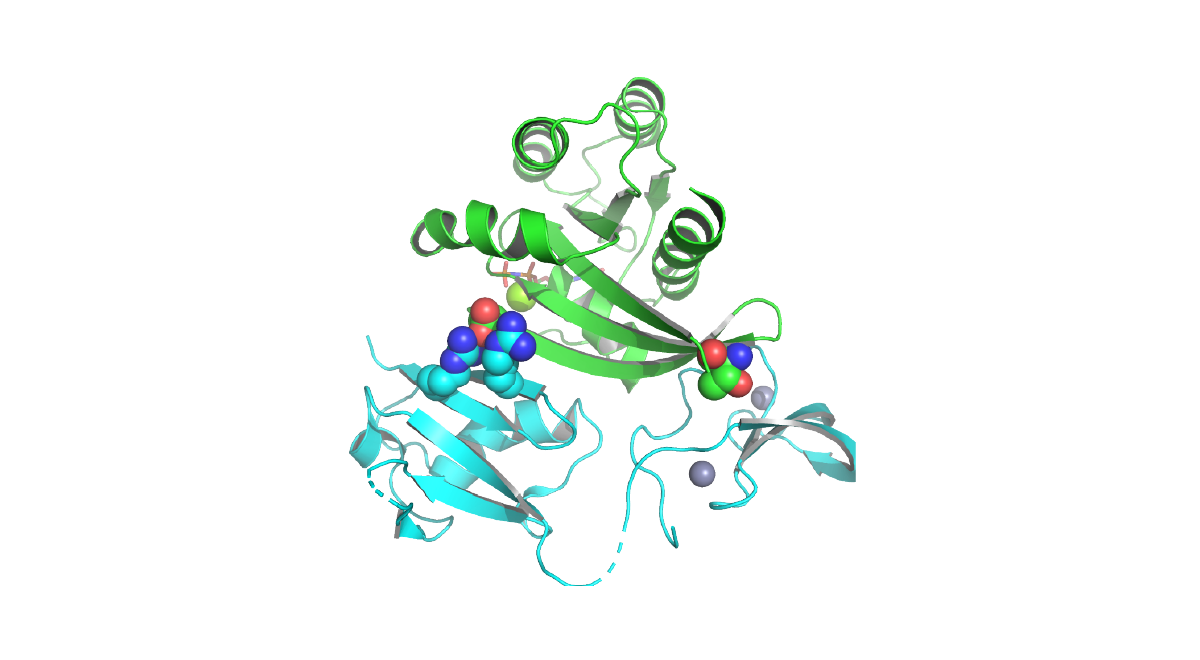
Codons 12, 13, and 61 of RAS are the most common targets of oncogenic mutations in cancer. We generated a novel “switchable” inducible Ras cell line system and we are leveraging it to investigate new therapeutic approaches to targeting oncogenic Ras. In collaboration with Dr. Kevin Shannon, we are targeting the palmitoylation-depalmitoylation cycle in NRAS mutant cancers, a process that is required for the transformation by oncogenic N-Ras. Because the normal K-Ras4b isoform does not require palmitate turnover, this approach has the potential to be selective for cancer cells that are dependent on oncogenic N-Ras. In collaboration with Dr. Kevan Shokat, we have also contributed to efforts to show that many Ras, Rho, and Rab GTPases exhibit targetable switch II pockets similar to one occupied by K-Ras(G12C) inhibitors. Notable differences in key residues offer potential for the development of optimized inhibitors for many members of this previously undruggable family.
Molecular Risk in Acute Myeloid Leukemia
The use of hematopoietic stem cell transplantation (HSCT) or bone marrow transplantation (BMT) as consolidation therapy for AML diagnosed in children, adolescents, and young adults has been debated for decades. We combined clinical, laboratory, and high-throughput sequencing data from the last two frontline AML clinical trials through the Children’s Oncology Group. Our study includes the largest contemporary pediatric AML BMT study cohort and is the first study to incorporate cryptic high-risk fusions that are only identified using RNA high-throughput sequencing methodologies. The results of our analysis are the first to indicate that children, adolescents, and young adults with high-risk AML defined by contemporary criteria benefit from HSCT or BMT in first complete remission.
Molecular Platforms for Detecting Residual Disease in Acute Myeloid Leukemia
Acute myeloid leukemia (AML) is an aggressive hematologic cancer associated with poor outcomes and current therapies are toxic. Therefore, distinguishing who will be cured with chemotherapy alone from those who require more intensive therapies is critical to improving cure rates in AML. The presence of small numbers of persisting leukemia cells after chemotherapy has become an important predictor of leukemia relapse. However, current assays used to detect residual leukemia have limited sensitivity and many patients with no detectable leukemia still go on to relapse. This underscores the need to identify and develop more accurate and sensitive leukemia detection assays for AML. We are developing novel platforms that harnesses best in class technologies to enable ultrasensitive detection of leukemia cells. We are validating these assays in collaboration with the Children's Oncology Group (COG), which will then fill a critical unmet need in the field of AML.
Integrated Transcriptomics and Proteomics to Identify Targets in AML
Acute myeloid leukemia (AML) remains a therapeutic challenge and survival outcomes for AML have not significantly improved for nearly 30 years due to a paucity of effective biologically based therapies. While immunotherapies targeting CD19 have yielded remarkable outcomes in B-cell acute lymphoblastic leukemia, identifying similar targets in AML remains a challenge due to overlapping immunophenotypes between leukemia cells and normal hematopoietic stem and progenitor cells (HSPCs) and transcriptional heterogeneity across AML subtypes. Therefore, in collaboration with Dr. Soheil Meshinchi (Fred Hutch), we integrated large transcriptome and proteome datasets from AML and normal tissues to identify potential targets expressed in leukemias, but not in normal bone marrow and other tissue types. Using this integrated approach, we identified several novel candidate immunotherapy targets. Importantly, a subset of these targets have clinically available antibody drug conjugates that are either FDA approved and/or undergoing early clinical development for other tumor types, serving as early candidates that we are currently pursuing with preclinical validation studies.